|
|
一:Kmeans算法基本思想:
k-means算法是一种很常见的聚类算法,它的基本思想是:通过迭代寻找k个聚类的一种划分方案,使得用这k个聚类的均值来代表相应各类样本时所得的总体误差最小。
k-means算法的基础是最小误差平方和准则。其代价函数是:
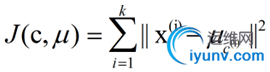
式中,μc(i)表示第i个聚类的均值。我们希望代价函数最小,直观的来说,各类内的样本越相似,其与该类均值间的误差平方越小,对所有类所得到的误差平方求和,即可验证分为k类时,各聚类是否是最优的。上式的代价函数无法用解析的方法最小化,只能有迭代的方法。k-means算法是将样本聚类成 k个簇(cluster),其中k是用户给定的,其求解过程非常直观简单,具体算法描述如下:
1、随机选取 k个聚类质心点
2、重复下面过程直到收敛 {
对于每一个样例 i,计算其应该属于的类:
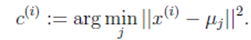
对于每一个类 j,重新计算该类的质心:
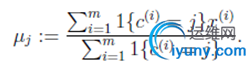
}
其伪代码如下:
********************************************************************
创建k个点作为初始的质心点(随机选择)
当任意一个点的簇分配结果发生改变时
对数据集中的每一个数据点
对每一个质心
计算质心与数据点的距离
将数据点分配到距离最近的簇
对每一个簇,计算簇中所有点的均值,并将均值作为质心
********************************************************************
代码如下:kmeans.py
from numpy import *
import time
import matplotlib.pyplot as plt
# calculate Euclidean distance
def euclDistance(vector1, vector2):
return sqrt(sum(power(vector2 - vector1, 2)))
# init centroids with random samples
def initCentroids(dataSet, k):
numSamples, dim = dataSet.shape ##numSamples = dataSet.shape[0]
centroids = zeros((k, dim)) ##初始化centroids用于存储质心点的坐标
for i in range(k):
index = int(random.uniform(0, numSamples)) ##随机生成(0,80)之间的数作为索引
centroids[i, :] = dataSet[index, :] ##根据随机索引初始化质心
return centroids
# k-means cluster
def kmeans(dataSet, k):
numSamples = dataSet.shape[0]
# first column stores which cluster this sample belongs to,
# second column stores the error between this sample and its centroid
clusterAssment = mat(zeros((numSamples, 2)))
clusterChanged = True
## step 1: init centroids
centroids = initCentroids(dataSet, k)
while clusterChanged:
clusterChanged = False
## for each sample
for i in xrange(numSamples):
minDist = 100000.0
minIndex = 0
## for each centroid
## step 2: find the centroid who is closest
for j in range(k):
distance = euclDistance(centroids[j, :], dataSet[i, :])
if distance < minDist:
minDist = distance
minIndex = j
## step 3: update its cluster
clusterAssment[i, :] = minIndex, minDist**2
if clusterAssment[i, 0] != minIndex:
clusterChanged = True
## step 4: update centroids
for j in range(k):
pointsInCluster = dataSet[nonzero(clusterAssment[:, 0].A == j)[0]]
centroids[j, :] = mean(pointsInCluster, axis = 0)
print 'Congratulations, cluster complete!'
return centroids, clusterAssment
# show your cluster only available with 2-D data
def showCluster(dataSet, k, centroids, clusterAssment):
numSamples, dim = dataSet.shape
if dim != 2:
print "Sorry! I can not draw because the dimension of your data is not 2!"
return 1
mark = ['or', 'ob', 'og', 'ok', '^r', '+r', 'sr', 'dr', '<r', 'pr']
if k > len(mark):
print "Sorry! Your k is too large! please contact Zouxy"
return 1
# draw all samples
for i in xrange(numSamples):
markIndex = int(clusterAssment[i, 0])
plt.plot(dataSet[i, 0], dataSet[i, 1], mark[markIndex])
mark = ['Dr', 'Db', 'Dg', 'Dk', '^b', '+b', 'sb', 'db', '<b', 'pb']
# draw the centroids
for i in range(k):
plt.plot(centroids[i, 0], centroids[i, 1], mark, markersize = 12)
plt.show()
def test_kmeans():
## step 1: load data
print "step 1: load data..."
dataSet = []
fileIn = open('F:/eclipse/workspace/K_meansTest/testSet.txt')
for line in fileIn.readlines():
lineArr = line.strip().split('\t')
print lineArr
dataSet.append([float(lineArr[0]), float(lineArr[1])])
## step 2: clustering... ###!!!从此开始缩进错误,不应该在for循环里面,应该和for循环在同一个级别
print "step 2: clustering..."
dataSet = mat(dataSet)
k = 4
centroids, clusterAssment = kmeans(dataSet, k)
## step 3: show the result
print "step 3: show the result..."
showCluster(dataSet, k, centroids, clusterAssment)
test_kmeans.py
from numpy import *
import time
import matplotlib.pyplot as plt
import kmeans
kmeans.test_kmeans()
调试程序中遇到的问题:
(1)提示AttributeError:“matrxi” have no "append" attribute!
错误原因:程序中缩进错误,应该将数据全部加载到dataSet列表中,再将dataSet列表使用mat()函数转化为矩阵,列表具有append()方法,而矩阵不具有此方法。
运行结果:
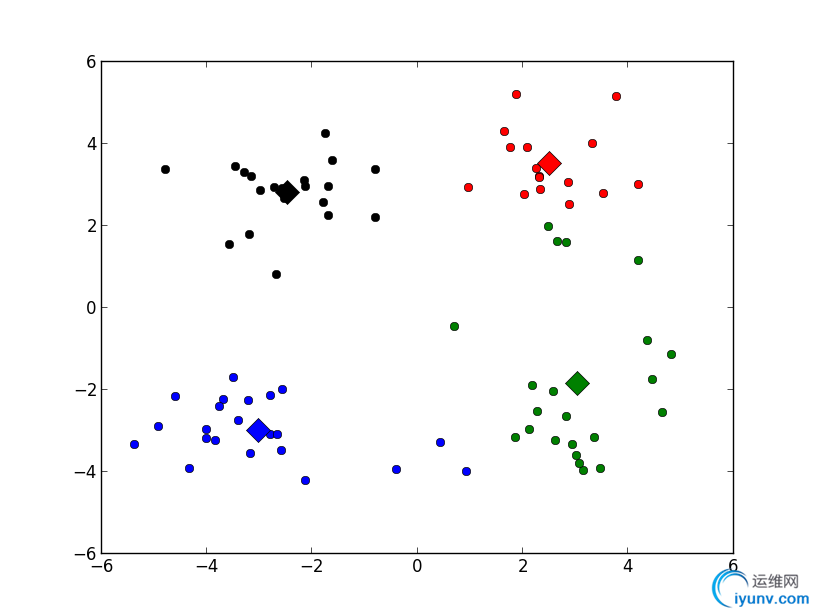
程序中相关知识注解:
mat()函数:数组转化为矩阵
random.uniform():函数原型为:random.uniform(a, b),用于生成一个指定范围内的随机符点数,两个参数其中一个是上限,一个是下限。
dataSet.shape:求取矩阵的形状
nonzeros(a):返回数组a中值不为零的元素的下标,它的返回值是一个长度为a.ndim(数组a的轴数)的元组,元组的每个元素都是一个整数数组,其值为非零元素的下标在对应轴上的值。
|
|